TrialPanorama: Developing Large Language Models for Clinical Research Using One Million Clinical Trials
Abstract
Developing artificial intelligence (AI) for clinical research requires a comprehensive data foundation that supports model training and rigorous evaluation. Here, we introduce TrialPanorama, a large-scale structured resource that aggregates 1.6M clinical trial records from fifteen global registries and links them with biomedical ontologies and associated literature. To demonstrate its utility, we build a pipeline that constructs 152K training and testing samples for eight key clinical research tasks. Three tasks support systematic review workflows, including study search, study screening, and evidence summarization. Five tasks focus on trial design and optimization, including arm design, eligibility criteria design, endpoint selection, sample size estimation, and trial completion assessment and rationalization. Benchmarking cutting-edge large language models (LLMs) reveals that generic LLMs have limited capability in clinical reasoning. In contrast, an 8B LLM we developed on TrialPanorama using supervised finetuning and reinforcement learning wins over the 70B generic counterparts in all eight tasks, with a relative improvement of 73.7%, 67.6%, 38.4%, 37.8%, 26.5%, 20.7%, 20.0%, 18.1%, and 5.2%, respectively. We envision TrialPanorama provides a solid foundation for future scaling of AI for clinical research.
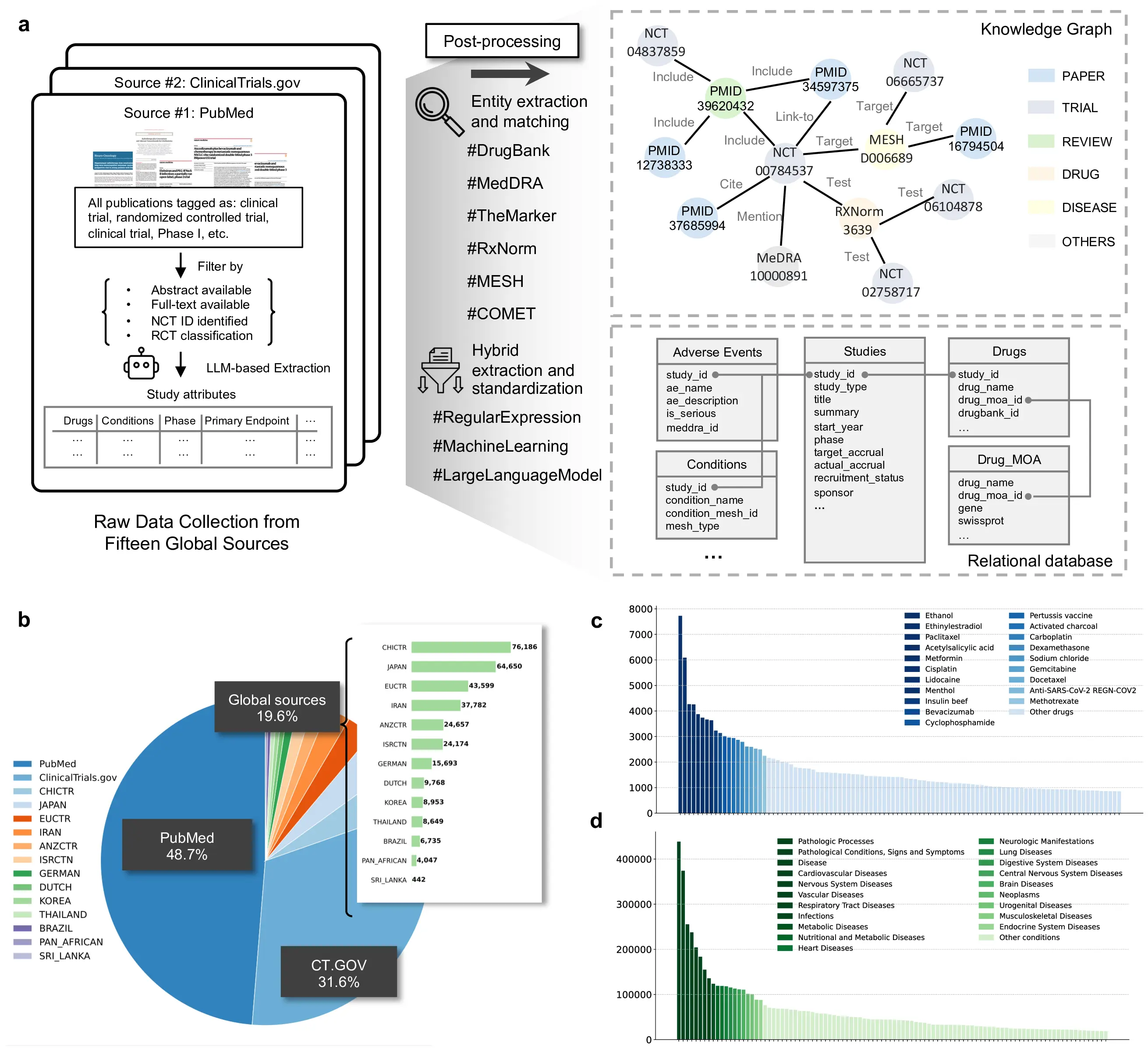
Figure 1: Overview of TrialPanorama and its end-to-end processing workflow. a, Raw clinical research data are collected from fifteen global sources, including PubMed, ClinicalTrials.gov, and international registries. LLM-based extraction, hybrid rule-based methods, and ontology-grounded normalization are applied to derive unified study attributes, followed by entity extraction and matching to DrugBank, MedDRA, RxNorm, MeSH, and other biomedical ontologies. The processed information is stored in both a relational schema and a heterogeneous knowledge graph linking trials, publications, drugs, diseases, and outcomes. b, Distribution of data sources in TrialPanorama showing contributions from PubMed, ClinicalTrials.gov, and aggregated global registries, together with a country-level breakdown of non-US registries. c, Most frequent drug entities mapped to standardized identifiers, illustrating the long tail distribution and coverage of therapeutic compounds. d, Most frequent disease and condition entities after ontology harmonization, demonstrating broad coverage across oncology, infectious disease, neurologic disease, metabolic disorders, cardiovascular disease, and other major therapeutic areas.
Database
The TrialPanorama database comprises ten main tables: studies, conditions, drugs, endpoints, disposition, relations, biomarkers, outcomes, adverse events, and results.
These tables are organized into four conceptual clusters:
- Trial attributes: Captures core metadata such as study title, brief summary, sponsor, start year, and recruitment status.
- Trial protocols: Describes the setup and design of the study, including tested drugs, targeted conditions, and patient group allocations.
- Trial results: Encompasses reported findings, including the trial outcomes, adverse events, and efficacy results.
- Study-level links: Encodes relationships across studies, such as mappings between registry records and corresponding publications, or reviews aggregating multiple studies on a common clinical topic.
Benchmark tasks
The TrialPanorama benchmark contains eight tasks across two categories: Systematic Review Tasks and Trial Design Tasks.
- Study Search: Retrieve relevant clinical trials based on a research question
- Study Screening: Determine if a study meets specific inclusion criteria
- Evidence Summarization: Synthesize findings across multiple related studies
- Arm Design: Design intervention and control arms with appropriate dosages
- Eligibility Criteria: Define inclusion and exclusion criteria for trial participants
- Endpoint Selection: Determine appropriate primary and secondary endpoints
- Sample Size Estimation: Calculate required participant numbers for statistical power
- Trial Completion Assessment: Predict likelihood of successful trial completion
Each task is designed to evaluate AI capabilities in supporting different aspects of clinical trial research and design.
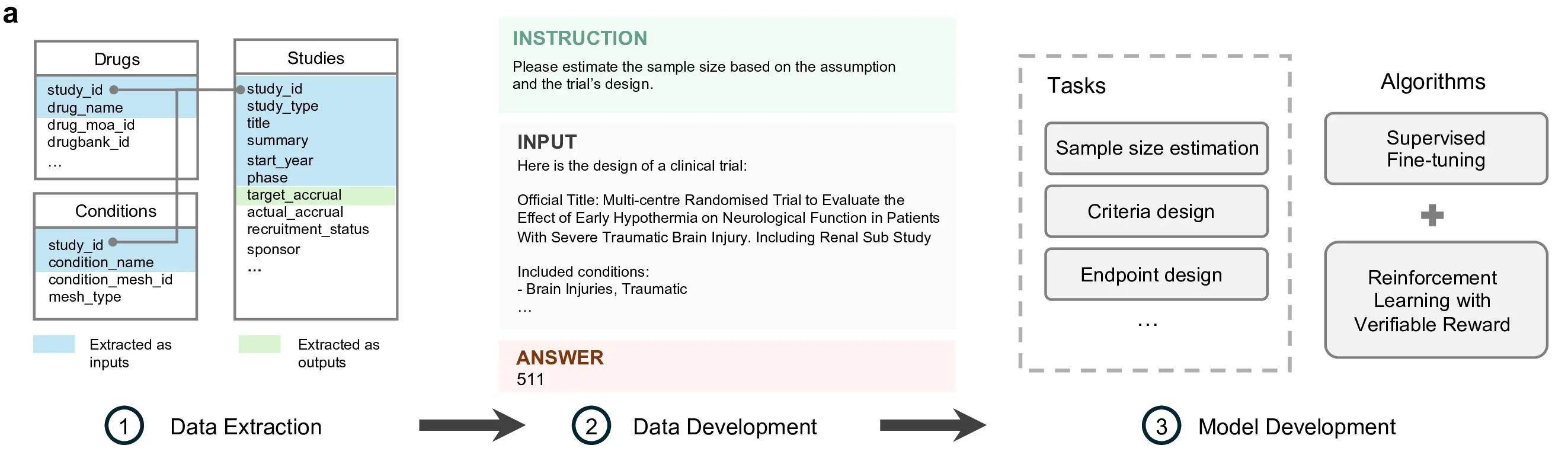
Figure 2: End-to-end workflow for transforming structured and semi-structured clinical trial records into supervised training examples. Extracted study attributes are composed into standardized instruction input answer triples, enabling the development of models for tasks such as sample size estimation, criteria design, and endpoint design.
Benchmark results
We evaluated cutting-edge large language models on the full suite of eight benchmark tasks spanning systematic review and clinical trial design workflows. Figure 3 presents a comprehensive overview of the benchmark structure (panels b and d) and model performance across all tasks (panel c), revealing substantial variation in capabilities and highlighting areas where domain-adapted models outperform generic counterparts.
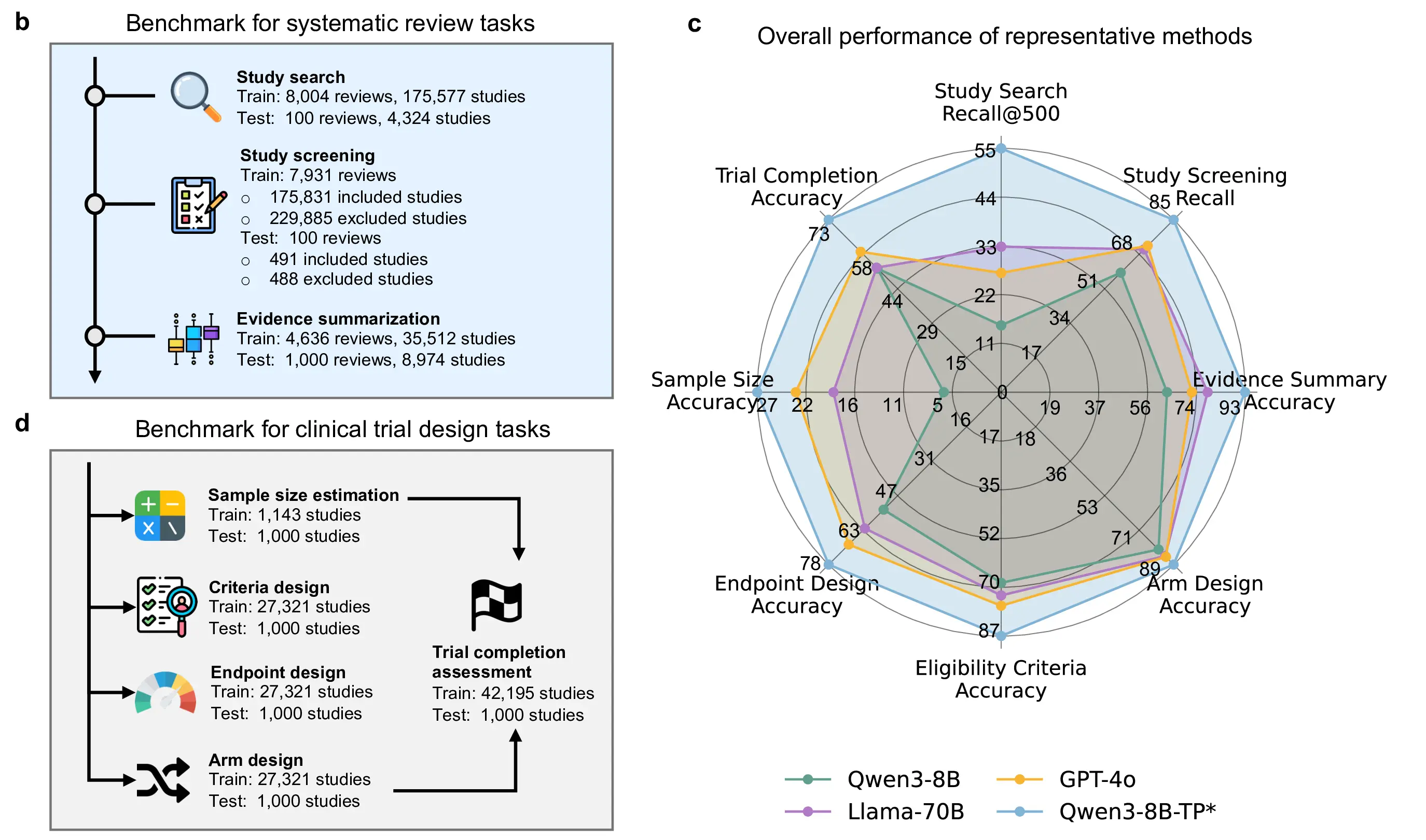
Figure 3: Benchmark overview and comprehensive evaluation results.b, Overview of systematic review tasks and dataset scale. c, Overall performance of representative LLMs across review and trial design benchmarks. d, Clinical trial design tasks and corresponding training and evaluation set sizes.
We benchmark cutting-edge large language models on eight core tasks spanning systematic review and clinical trial design (Figure 3b–d). These tasks are constructed directly from real-world clinical trial records and review corpora, and are designed to evaluate clinical reasoning, trial design competence, and evidence synthesis capabilities. The benchmark includes tasks ranging from literature-level reasoning to structured trial design decisions, providing a comprehensive assessment of LLM performance in clinical research settings.
Across all eight tasks, generic large language models exhibit limited capability in clinical reasoning (Figure 3c). Despite strong general language understanding, even large-scale models with up to 70B parameters struggle to reliably solve clinically grounded tasks, particularly those requiring domain-specific knowledge, structured trial reasoning, or integration of multiple clinical constraints. These results indicate that model scale alone is insufficient for robust performance in clinical research benchmarks.
In contrast, Qwen3-8B-TP*, an 8B-parameter model adapted on TrialPanorama using a combination of supervised fine-tuning and reinforcement learning with verifiable reward, outperforms all generic counterparts across every benchmark task. Despite being nearly an order of magnitude smaller than the largest evaluated models, Qwen3-8B-TP* achieves consistent and substantial gains, with relative improvements of 73.7%, 67.6%, 38.4%, 37.8%, 26.5%, 20.7%, 20.0%, 18.1%, and 5.2% across the eight tasks.
These results demonstrate that domain-aligned training is a critical factor for enabling LLMs to perform clinical research tasks. A carefully adapted 8B model trained on high-quality, task-aligned clinical trial data not only closes the gap with much larger generic models, but consistently surpasses them across all evaluated tasks. This highlights the importance of domain-specific post-training over brute-force scaling for deploying LLMs in clinical research.
Reference
Please kindly cite our paper if you use our code or results:@article{wang2025trialpanorama,
title = {Developing Large Language Models for Clinical Research Using One Million Clinical Trials},
author = {Wang, Zifeng and Lin, Jiacheng and Jin, Qiao and Gao, Junyi and Pradeepkumar, Jathurshan and Jiang, Pengcheng and Lu, Zhiyong and Sun, Jimeng},
year = {2025},
}