January 20, 2026 · Zifeng Wang, PhD, Keiji AI
Connecting the dots in biomedical research is a persistent headache. Anyone who has tried to link a specific gene mutation found in a PubMed abstract to its corresponding signaling pathway in KEGG, and then to a druggable compound in ChEMBL, knows exactly what I mean.
Yes, the new wave of "Deep Research" agents, like those from OpenAI, Google, and Perplexity, has shown that AI can browse the open web and synthesize information. But for serious biomedical discovery, browsing the web isn't enough. We don't just need summaries of internet-scale text; we need to systematically navigate heterogeneous Knowledge Graphs (KGs). These sources, spanning literature, clinical trials, pathways, and chemical annotations, come with structural differences and limited alignment that generic LLMs simply cannot bridge out of the box.
This is where we saw the next frontier for AI scientists. If generic AI is a librarian, we wanted to build a Deep Research Agent that acts like a specialized investigator. This is DeepEvidence.
From "Browsing" to "Systematic Exploration"
Most existing AI agents perform "shallow exploration" — they issue a few searches and call it a day. DeepEvidence changes the procedure by explicitly building an internal Evidence Graph as its persistent memory. As the agent traverses different databases, it incrementally records entities (genes, drugs, diseases) and their relations, providing a structured, auditable record of the discovery process.
To make this exploration truly "deep," we designed an orchestrator that coordinates two specialized sub-agents:
- Breadth-First ReSearch (BFRS): Think of this as the "radar." It issues broad, multi-graph queries to rapidly survey first-hop neighborhoods, capturing a wide set of candidate targets across 17+ different biomedical APIs.
- Depth-First ReSearch (DFRS): This is the "detective." It builds on the leads found by BFRS, recursively tracing references and citations to uncover complex mechanisms hidden deep in the literature.
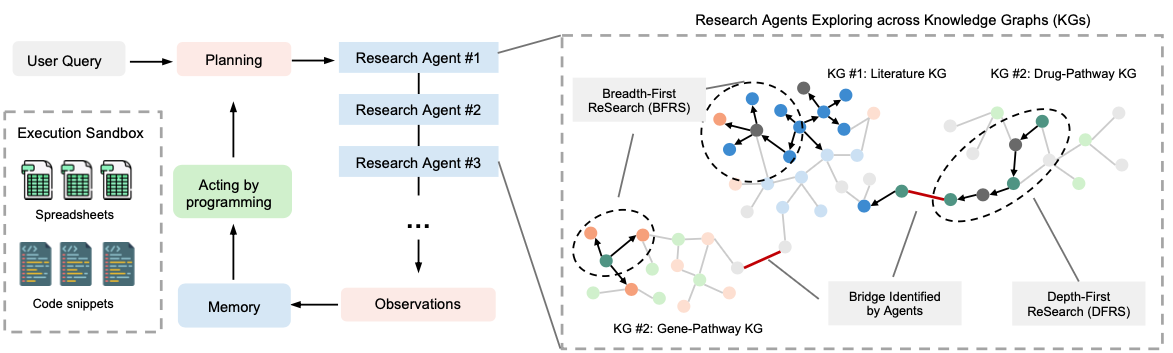
The Secret Sauce: The Execution Sandbox
In my previous post, I talked about how vertical, domain-specific data is crucial. In DeepEvidence, we take that a step further with an Execution Sandbox.
Instead of just generating text, our agents write and execute Python code to conduct research programmatically. When an agent needs to fetch data from OpenTargets or analyze a list of 200 clinical trials, it writes a script to query the APIs, persists the results in spreadsheets, and performs downstream data analysis. This ensures that the research workflow is not just a "black box" of text generation, but a reproducible and efficient scientific procedure.
Performance: Bridging the "Reasoning Gap"
We put DeepEvidence to the test on humanity's toughest medical benchmarks, and the results were, frankly, a breakthrough.
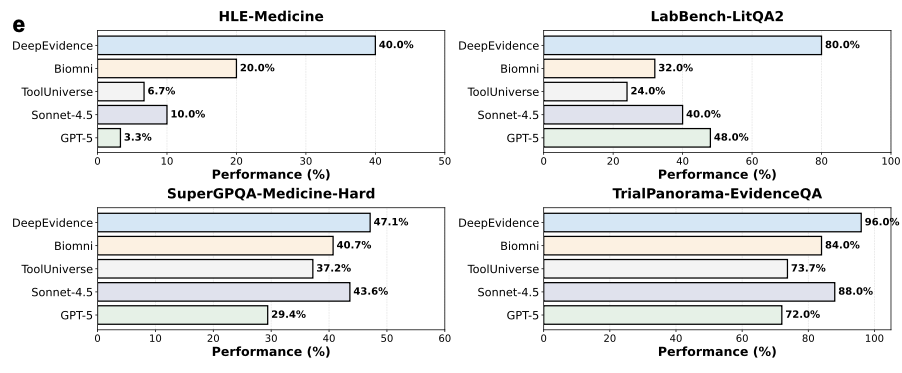
On HLE-Medicine (Humanity's Last Exam) — a test designed to be the "last exam" for AI — DeepEvidence achieved an accuracy of 40.0%. For context, the frontier models like GPT-5 and Sonnet-4.5 equipped with standard search capabilities only hit 3.3%.
Even when compared to other specialized biomedical agents like Biomni (20.0%) or ToolUniverse (10.0%), DeepEvidence's structured exploration and Evidence Graph memory allowed it to more than double their performance on complex reasoning tasks.
Real-World Discovery
We've already seen DeepEvidence handle tasks across the full drug development lifecycle:
- Drug Discovery: Identifying high-confidence therapeutic targets by parsing pathway topology.
- Clinical Trials: Estimating sample sizes and designing drug regimens by reviewing historical dosing patterns.
- Evidence-Based Medicine: Detecting "evidence gaps" in systematic reviews with a 90% detection rate, compared to just 10% for generic PubMed-search LLMs.
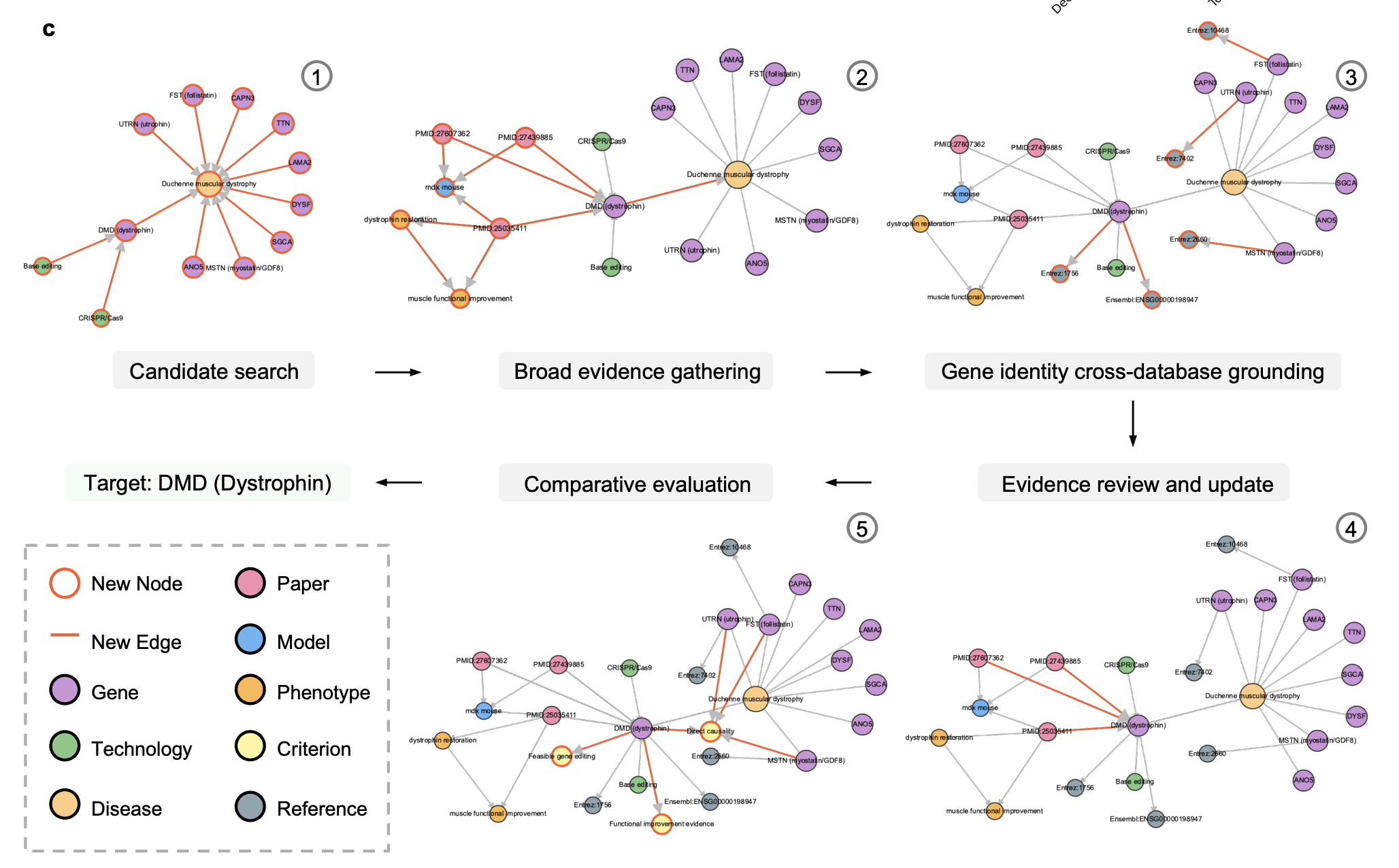
Looking Forward
DeepEvidence represents a shift from AI that "recites" facts to AI that "investigates" them. By grounding LLM reasoning in the structured reality of Knowledge Graphs, we are moving closer to a world where AI agents can assist human experts in generating and validating scientifically meaningful hypotheses through a transparent, deep research paradigm.
We've made the code and datasets public — because the only way to accelerate discovery is to let deep research with biomedical knowledge bases become an easier job. DeepEvidence is ready to use in Keiji AI's TrialMind platform for various biomedical research applications.
References & Resources
Paper: DeepEvidence: Empowering Biomedical Discovery with Deep Knowledge Graph Research
Code: BioDSA / DeepEvidence
Data: Hugging Face Datasets